I’ve been asked more and more for hints and best practices when working with R. It can be a daunting task, depending on how deep or specialised you want to be. So I tried to keep it as balanced as I could and mentioned point that definitely helped me in the last couple of years. Finally, there’s lots (and I mean, LOTS) of good advice out there that you should definitely check out - see some examples in the Quick Reference section below.
01. Use R projects. Always.
Human Civilization was built on conventions. R scripts have them, too. For example > Any resident R script is written assuming that it will be run from a fresh R process with working directory set to the project directory. It creates everything it needs, in its own workspace or folder, and it touches nothing it did not create.
You want your project to work. Not only now, but also in 5 years, even if folder and file paths have changed. Also, you want it to work when your collaborator runs it from their computer. Projects create convention that make it possible.
Basically, they create the environment where
- all code and outputs are stored in one set location (no more
setwd()!) - relative file paths are created - this guarantees better reproducibility
- clean R environment is created every time you open it (no more
rm(list = ls())!)
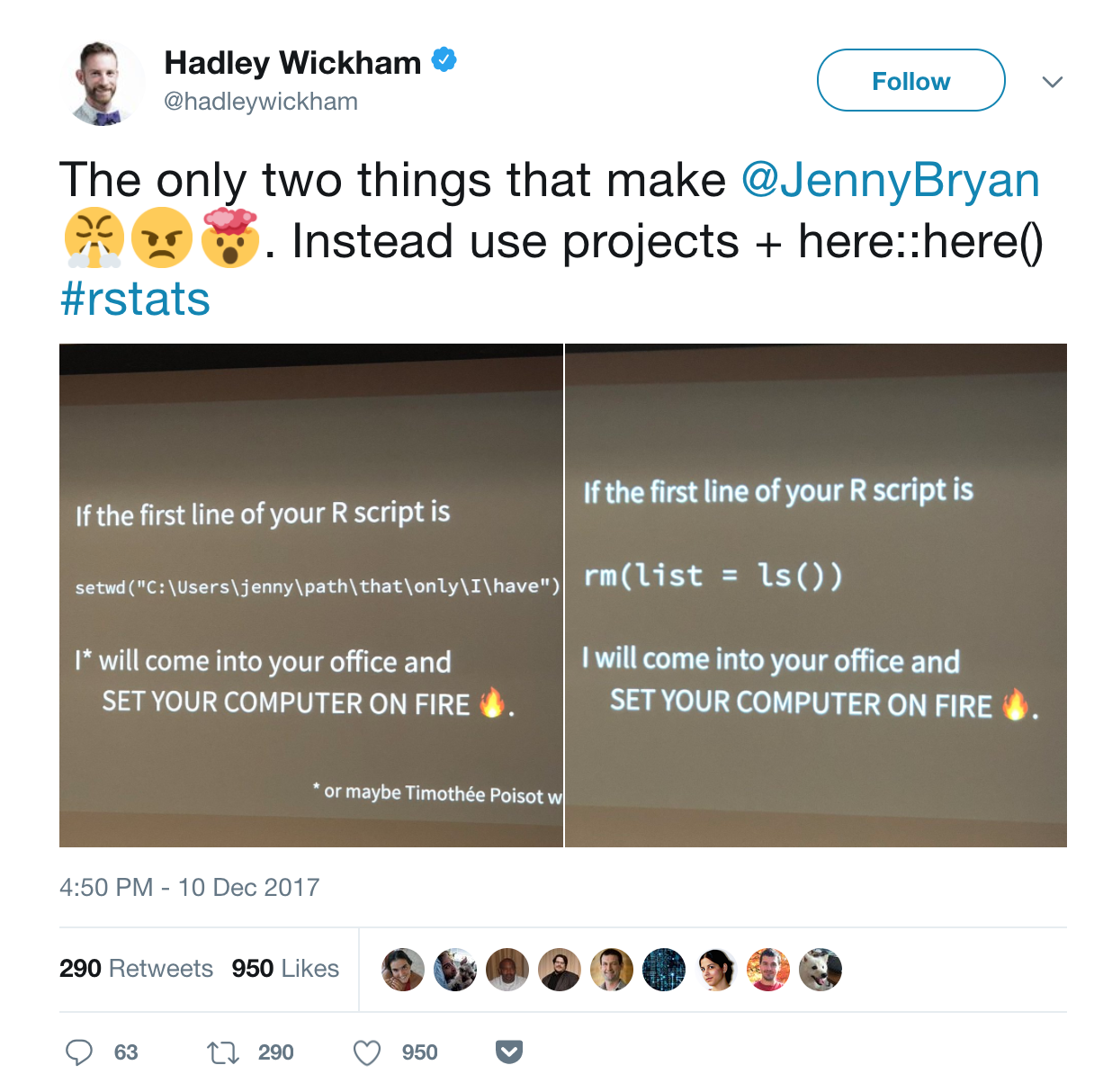
If you’re not convinced, read a more thorough explanation given by Jenny Bryan.
So, go on and start a New Project today! Otherwise, your computer may be at risk of being set on fire…
02. Describe the purpose of your code / project.
Before you even load the packages or import the data, state the purpose and content of your script in hashed code.
### Data import and cleaning from sources A, B and C.
### This script is part of Segmentation project.
Naturally, you can update it along the way, but having such description in place from day one will guarantee that whoever has access to your script will instantly know what he/she is looking at. Also, if you leave it for later, the chances are that you’ll forget to do it altogether and believe me, in X months time when you go back to your code it won’t be that clear what were you trying to do here.
03. Load all necessary packages in the beginning of your script & consider calling functions in a package via ::.
Ok, I know how it is: you load some basic packages to start with, but then the analysis takes to wondrous and wonderful places that require more specialized libraries. So you install and load them along the way… WRONG! Whenever you have to load a new package, go back to the beginning of your script and load them there. This way whenever you (or someone else) have to run the script again, they will have ALL necessary libraries to run it and won’t get stuck somewhere in the middle of the execution because the function they called is not recognized. It’s coding. Small things matter.
Also, if you don’t want to load the whole package just for the sake of using a single function, consider specifying package directly via ::. Use this text cleaning exercise as an example:
library(readr)
library(dplyr)
tweets <- read_csv("tweets.csv")
all_tweets <- tweets %>%
rename(author = handle) %>%
select(author, text) %>%
mutate(text = qdapRegex::rm_url(text)) %>% #removes URLs from text
na.omit()
Here, I’m expecting to use readr and dplyr packages regularly and I’m using qdapRegex library only once - to remove URL’s from my tweet data. In this case I call rm_url() once and I specify the package that it comes from. More about in here. Easy peasy.
!!! UPDATE !!!
As Brent Auble correctly pointed out, if you decide to use :: way of calling the package, make sure to put the comment clarifying that you’re going to use this package - preferrably in the load section of the script - so that the user installs the package before running the script.
Also, Maëlle Salmon made me realise that there’s already an excellent package packup that organises your code so that any mentioned libraries anywhere in the script automatically end up in the beginning of your code. Genius!
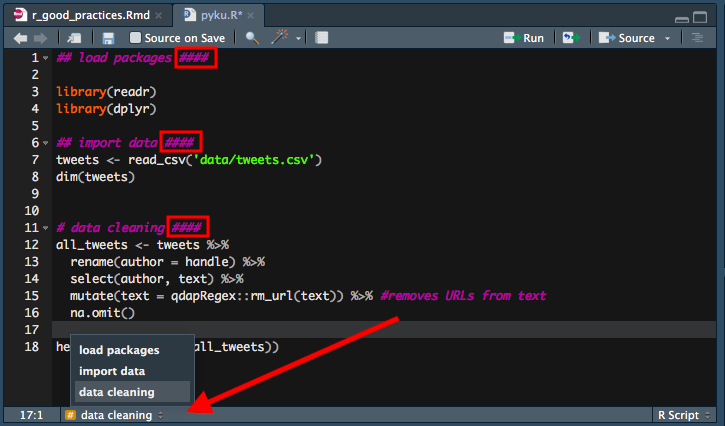
04. Name your code sections and use them for quick navigation.
Your code will grow. Sometimes it will turn into an R script equivalent of the Bible, at least in terms of volume. To keep it organized, keep your code nice and tidy by assigning relevant code chunks to different sections that can be later folded/unfolded; you can also easily navigate yourself through chunks by using ‘drop-up’ menu at the bottom of the script screen.
You create a new code section by writing #### or ---- at the end of any comment that is to become a new code section.
See the image below for a simplified example:
!!! UPDATE !!!
There’s even a keyboard shortcut in RStudio for inserting code sections: Ctrl+Shift+R for Windows users and Command+Shift+R for Mac users. Even Hadley Wickham can tell you that!
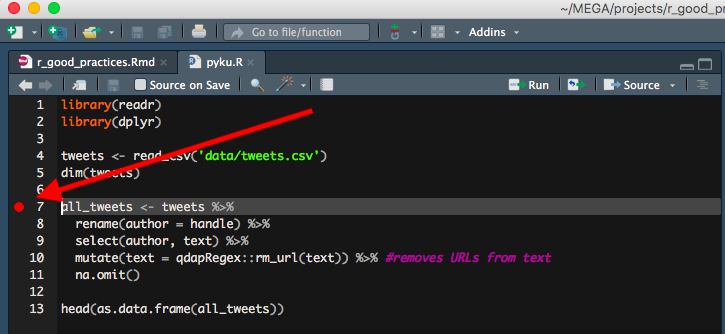
05. Make your life easier and mark your code when you need to.
Did you know that if you click anywhere left from line numbers in RStudio, it will create a red mark? I think this is one of the most underused features in RStudio, and a very useful one, too! Imagine having to define a function using another part of a very long R script? Instead of finding a relevant part of the script and then spending seconds or minutes on finding the function that you were about to define, you first mark that function, go to the relevant bit of code and then have no problems finding the function to be defined again, as the red mark will tell you where to look. BTW, it works only in already saved R scripts and it doesn’t work in RMarkdown files.
!!! UPDATE !!!
I didn’t know at the time of writing, but as Jonathan Carroll and many after him pointed out: the red dot actually indicates a debug point. If you leave it in the script while running the code, it will start a debugging process and therefore stop executing your code. Therefore always remember to remove the red dot when you run/source your code!!. Still, I find it pretty useful for temporary marking purposes ;)
06. Write your code as if someone was going to use it without communicating with you. From. Day. One.
I know, I know. We all mean to do it, but we rarely do. I always promise myself that I’ll be thoroughly commenting my code from the very beginning of the project but even these days I find it difficult to do it consistently. Why? Because:
- analysis itself is more important (I’m telling myself).
- I know what I’m doing.
- I (usually) have no direct collaborators that are using the code.
But how short-sighted those arguments are! The reality is that
-
even most precious and relevant piece of analysis is useless if you or others don’t understand it (more about it below)
-
you know what you’re doing at the moment, but it won’t feel the same way in a month or two when you have moved on to another project but someone asked you an innocent question about how you defined that key variable… Our memory is fallible. Make sure you don’t have to rely on it with every single piece of code you produce
-
even you don’t have active collaborators at the time of doing analysis, someone will have to use your code sooner or later. You’re not going to be in the same position for the rest of your life. You’re creating legacy that ONE DAY someone will use, no matter how far away it seems right now.
What makes good, reproducible code?
- generous and clear comments
- logical and efficient code
- code that is ideally timed and tested
!!! UPDATE !!!
I had discussions online and offline whether your code alone can be used as sufficient documentation. The argument is that clean and clear code should stand for itself, no comments are necessary. There was a time when I thought that, too, but as Mark Sellors pointed out: any code will explain what you did, but it will rarely say WHY you did it. So comment your code generously, please!
07. Name your files like a Pro.
Jenny Bryan is probably the first very high profile R-user I know who’s been actively highlighting the importance of things that not many people talk about: file naming is one of them. I’m not going to reinvent the excellent wheel that she has already invented, so I’m only going to summarize her advice here. But for more detail, please, please, have a look at Jenny’s slides.
So, what do all good file names have in common? They are:
**MACHINE READABLE **
This means that file names are
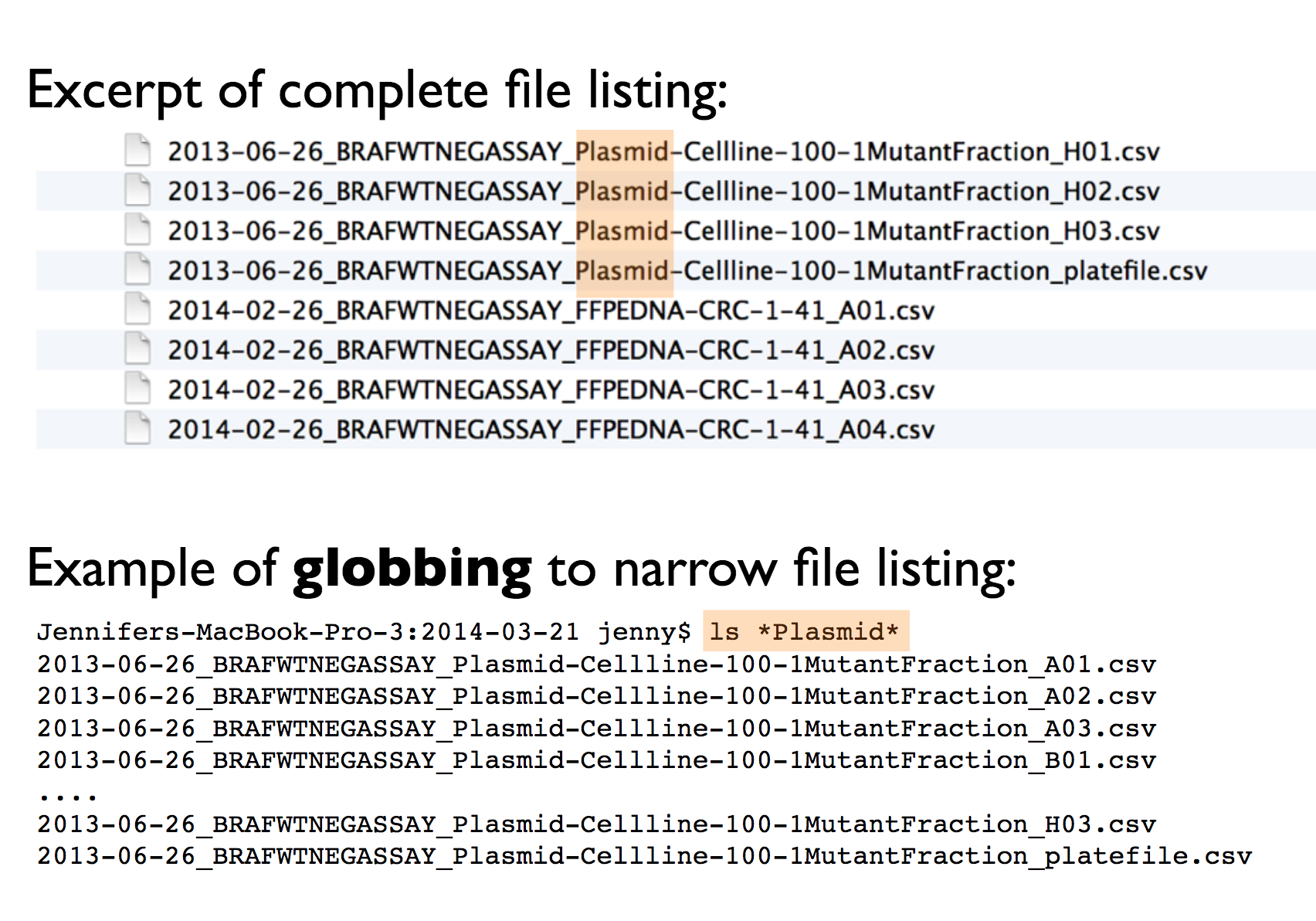
- regular expression and globbing friendly. Meaning? You can search them using key words (with
regexand/orstringrpackage). In order to make that easier/possible, remember to avoid spaces, punctuation, accented characters, case sensitivity.
This way it will be easy to search for files later and/or narrow file lists based on names.
Jenny’s example:
- easy to compute on using delimiters: this means that the file names should have a consistent name structure, where each section of the name has its role and is separated from other name sections with delimiters. This way it will be easy to extract information from file names, e.g. by splitting.
See Jenny’s example below for clarification:

HUMAN READABLE
Human readable means exactly what is says on the tin:
Easy to figure out what the heck something is, based on its name, as simple as that :)
I should add, it’s easy to figure it out also for someone who doesn’t know your work very well (this is a clear reminder of the previous point). I love the example below:

THEY PLAY WELL WITH DEFAULT ORDERING
The reality is that your computer will sort your files for you, no matter whether you like/need it or not. Therefore:
-
put something numeric in your file name first - if the order of sourcing files doesn’t matter, stating when the file was created is always useful. Otherwise, you can indicate the logical order of the files (I’ll come back to that in point 9).
-
use the
YYYY-MM-DDformat for dates (it’s ISO 8601 standard!) EVEN when you’re American :) -
left pad other numbers with zeroes - otherwise you’ll end up with
10before1and so on.
Again, one of Jenny’s examples:

I couldn’t believe how much these little hacks improved the flow of my work. Paying attention to file names was also appreciated by my coworkers, I’m sure.
08. If you have to copy/paste excerpt of code 3 or more times, write a function for it.
.. as Hadley Wickham said many many times in his and Charlotte’s course on DataCamp. It doesn’t only teach you to write more elegant and efficient code, but also it makes it more readable for yourself and others.
To learn how to write good functions, you can have a sneak peak into the first chapter of the DataCamp course for free (the rest is available under paid subscription) or read this DataCamp tutorial.
09. With big, complex data projects use project pipeline.
I’m not sure if project pipeline is an official name for what I want to talk about, but for a sake of the argument, let’s call it a project pipeline ;) Namely, sometimes running a full project from one script - with even clearest and informatively named code sections - is simply difficult, if not unfeasible. Particularly when big(ish) data is involved: imagine trying to run a hefty model on data that requires importing and cleaning, where import into R alone takes about an hour (real life scenario!). You wouldn’t possibly want to do it every time you open the project, right?
So what you do instead is write one script for data import and save the data.frame with useful data as an .RData file (using, e.g. save(data1, file = "data.RData"). Then start a new script for, let’s say, data cleaning where you load previously imported data.frame from the .RData file (using load("data.RData")). Then you clean the data and save it as yet another .RData file. Then you write the third script where you load the clean data.frame from the second .RData file and you use it to run your model. Jenny Bryan’s advice on file naming comes in handy here, as you want to name and order your scripts or their outputs logically to avoid surprises in the future.
This way you create a clear structure of building blocks, as well inputs and outputs. Additionally, once the data has been imported and cleaned, you can jump straight away into next steps of analysis/modelling without wasting any time on the first two steps.
I started using this approach in big and complex projects after reading this SO answer and I never looked back.
!!! UPDATE !!!
Lots of comments regarding this one.
First, I didn’t mention it explicitly, but it mentioned in the Stack Overflow reply I refer to: if you define a number of functions, it’s a good practice to define them in a separate script that you can later source. Blazko is clearly a fan of this solution :)
Secondly, saving your data objects as .rds file is probably a better solution than using .RData, as you can directly assing it to another object in a new session (right, Kees Mulder ?) or assign a bunch of similar object into a list (wink wink, Martin Johnsson ). It’s also an official recommendation of Jenny Bryan.
Finally, if you need a reproducible way of running a number of connected script and/or visualising how they are connected, search no more: drake package is here to help! Really impressive tool, had no idea it existed at the time of writing this post, so thanks Peter Higgins for telling me about it.
10. Never save your workspace.
Again, there were others that already said it before me and said it much better than I would have. So there you go:
Loading a saved workspace turns your R script from a program, where everything happens logically according to the plan that is the code, to something akin to a cardboard box taken down from the attic, full of assorted pages and notebooks that may or may not be what they seem to be. You end up having to put an inordinate trust in your old self. I don’t know about your old selves, dear reader, but if they are anything like mine, don’t save your workspace.
For details, see Martin Johnsson’s excellent blog post that I quoted above.
11. Before publishing/sharing your code, run it in the fresh workspace.
This almost goes without saying, but if I got a £ for every time I forgot to do it… Basically, stuff happens when you write your code: you work with multiple files, maybe you loaded a package in one of them but forgot to do it in the rest of them? If that’s the case, you’ll be able to run all scripts in this particular session but if you send someone the code without the imported function… they will fail to run it. The same will happen if you try to run the script on its own, on your own machine.
Anyway, this doesn’t need much explaining. Simply make sure that you re-run your code in a fresh session before you take it further. Amen.
That’s it for now.
So.. these are my ‘prime hints’ - if someone had told me about them 2 years ago, my #rstats life would have been much much easier. By no means is this list complete or exhaustive, but my intention was to highlight the points that I personally found most useful in my daily fun with R. Hope it will help someone!